Assessment of Genetic Variability in Long-lived Cupressus sempervirens var. horizontalis Using SSR Markers 
2. Faculty of Natural Resources, University of Gorgan
3. Faculty of Natural Resources, University of Tehran
4. Research Group of the Technology of Natural Sustainable Ecosystems
5. Resrach Institite of Forests and Rangelands
 Author
Author  Correspondence author
Correspondence author
Plant Gene and Trait, 2012, Vol. 3, No. 8 doi: 10.5376/pgt.2012.02.0008
Received: 12 May, 2012 Accepted: 20 Jun., 2012 Published: 28 Jun., 2012
Soudabeh et al., 2012, Assessment of Genetic Variability in Long-lived Cupressus sempervirens var. horizontalis Using SSR Markers, Plant Gene and Trait, Vol.3, No.8 43-49 (doi: 10.5376/pgt.2012.03.0008)
The ability of adaptation to environmental changes depends greatly on the genetic diversity of the species. As a member of Cupresacea, Cupressus sempervirens var. horizontalis is one of the four native conifer trees in Iran that distribute in different geographical provinces individually with the high longevity character. Assessing genetic diversity is considered vital for formulating conservation strategies of endangered species such as Cypress. SSR molecular markers were employed to assess genetic diversity. Nei’s gene diversity (HE) ranged from 0.16 to 0.32 and the average expected heterozygosity (HE) was 0.26. The mean Shannon indices (HO) was 0.41. The average Polymorphism Information Content was 0.26. Dendrogram was constructed using UPGMA method divided all individuals into 4 major groups. Principal coordinate analysis shows the first two components account for 51.87% of the total variation. The study implies that more variation needs to be introduced in the existing population for species persistence.
Background
Genetic diversity of endangered species has always enthused evolutionary and conservation biologists. The ability of a species to adapt to environmental changes depends greatly on the genetic diversity in the species (Neel and Ellstrand, 2003; Anand et al., 2004). As a member of Cupresacea, Cupressus sempervirens var. horizontalis is one of the four native conifer trees in Iran. This tree species have been sparsely distributed in different geographical provinces. It is native to the eastern Mediterranean region, in northeast Libya, southeast Greece (Crete, Rhodes), southern Turkey, Cyprus, Northern Egypt, western Syria, Lebanon, Palestine, Malta, Italy, western Jordan, and also a disjunctive population in Iran. The natural stands of this species in Iran distribute in Mediterranean regions. In addition, individuals of Cupressus sempervirens with the high longevity character distribute in all regions of Iran. These trees have a long age, so one of the most characteristic of these individuals is high resistance to biotic and abiotic stresses. C. sempervirens is a medicinal plant. The dried leaves of this plant are used as an emmenagogue and a remedy for the stomach pain (Castro, 1998) as well as for diabetes (Assadi, 1998). The dried fruit of this plant is used for inflammation treatment (Mascolo et al., 1987), toothache, laryngitis (Darias, 1989), as a contraceptive (Jochle, 1962), astringent, and antiphrastic (Ponce-Macotela et al., 1994). Genetic variations within and among populations of species are critically important for any conservation programs, because the long-term survival of such a species strictly depends on the maintenance of sufficient genetic variation to facilitate adaptations to long-term environmental changes (Gitzendanner and Soltis, 2000).
The analysis of the genetic variation within and among populations of the species is crucial for understanding their future maintenance and developing improvements in conservation programs (Mahar et al., 2011). Meanwhile, there are many available molecular methods to analyze the genetic variability in plant species, but no single method is universally applicable (Mahar et al., 2011). Many researchers use molecular methods to estimate genetic diversity (Su et al., 2009; Hafezi Shahroodian et al., 2011; Buiteveld et al., 2007; El-Kassaby et al., 2003; Wang et al., 2004).
In the present study, we used Simple Sequence Repeat (SSR or microsatellite) markers to estimate the genetic variability in C. sempervirence individuals. SSRs are tandem repeated units of nucleotides that are abundant in prokaryotic and eukaryotic genomes and are ubiquitously distributed in both genome’s protein-coding and non- coding regions (Field and Wills, 1996; Toth et al., 2000). The hyper variability and co-dominance of SSRs, their dispersion throughout genomes and suitability for automation are the principal reasons for their wide utility (Powell, 1996; Jarne, 1996; Gupta, 1996). These markers have been used to characterize genetic diversity in plants (Zhang et al., 2007; Hafezi Shahroodian et al., 2011; Chen et al., 2009; Angielina et al., 2011).
Sixty three individuals of Cupressus sempervirens from all regions of Iran were examined to quantify genetic diversity and genetic structure by SSR markers. These trees were selected for the research because of their adaptive potentials. The spatial variation in tree genetic diversity, in turn, determines the adaptability of tree populations to environmental change and is thus essential for the long-term sustainability of forest ecosystems (Giannini et al., 1991; Rehfeldt et al., 2002; Bilgen and Kaya, 2007; Savolainen et al., 2007). The main objective of the present study was to evaluate genetic variability and degree of genetic divergence within long-lived individuals of Cupressus sempervirence.
1 Results
The total number of alleles per locus is listed in Table 1. A total of 113 alleles were detected, the mean number of alleles per locus was 14.12. The number of observed alleles in all the individuals per locus varied from 8 for Cyp84 and Cyp258 to 25 for Cyp250. The most of eight loci assayed possessed a high level of polymorphism, with the number of the effective alleles per locus ranging from 1.23 at Cyp250 to 1.55 at Cyp293. The PIC value was also estimated, as shown in Table 1 with the highest value (0.32) for Cyp52 and the lowest values (0.16) for Cyp250 (Figure 1). Nei’s gene diversity (HE) ranged from 0.16 to 0.32 and the average expected heterozygosity (HE) and the mean Shannon indices (HO) was 0.26 and 0.41, respectively.
Table 1 Observed and effective number of alleles, Nei’s gene Diversity, Shannon’s index and polymorphic information content (PIC) for eight microsatellite loci analyzed in 63 individuals of Cupressus sempervirence |
Figure 1 The high level of polymorphism resolved on 5% Polyacrylamide gel and Amplified DNA fragment generated by Cyp52 primers fluorescently visualized in GS2000 |
In order to distinguish the best clustering and similarity coefficient calculation methods, the cophenetic correlation, a measure of the correlation between the similarity represented on the dendrograms and the actual degree of similarity, was calculated for each method combination. Among different methods, the highest value (r=0.74) was observed for UPGMA clustering method based on Simple Mathing’s similarity coefficient (Table 2). Therefore, the dendrogram was constructed based on this method was used for depicting genetic diversity of genotypes (Figure 2). Cluster analysis divided the 63 individuals into four groups. Group 1 contains two individuals as Pirdal 1 and 2 in Gilan province which was placed in a separated cluster with a very low similarity to other groups. Group 3 contains eleven individuals in two provinces Gilan and Fars including Poshtehan, Poshtehan shahed 1, 2, 4 and Tange kherghe 1, 3, 4, 5, 6 and Tange kherghe shahed 1, 2. Also group 4 contains two individuals as Tange soulak 3 and Sirch 1 in Kohgiloyeh and Kerman province, respectively. All other 48 individuals placed in group 2 belonged to 13 provinces in Iran.
Table 2 Comparison of different methods for constructing dendrogram |
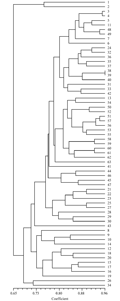 Figure 2 Dendrogram showing the genetic relationship among 63 individuals of C. sempervirence based on SSR data, UPGMA clustering method and Simple Mathing's coefficient. 63 individuals were divided in 4 different clusters |
Principal coordinate analysis (PCo) based on genetic similarity metrics was used to visualize the genetic relationship among individuals. The first two eigenvectors accounted for 51.87% of the total molecular variation. Therefore, PCo results confirmed the results of cluster analysis (Figure 3) except that Pirdal 1 at group 1 which is mixed to group 3 and also Sirach1 at group 4 which is mixed to group 2. The genetic distances among studied individuals were represented in Table 3. The highest genetic distance was recorded between Pirdal 2 in Gilan and Dorbid 1 in Yazd and lowest one between Dehbakri 1 and 2 in Kerman province. The individuals with the lowest genetic distance with Dehbakri 1 (Kerman P.) as a most diameter tree were Dehbakri 2 (Kerman P.) and Chevar 1 (Ilam P.) and the closest to Abarkooh as the most famous tree were Lar (Kohgilooyeh & Boi. P.) and Kashmar (Khorasan Razavi P.) as well.
 Figure 3 The scatter plot of the first and second principal coordinates analysis on 63 individuals of Cupressus sempervirence based on the SSR obtained with eight primer combinations |
 Table 3 Genetic distances among 63 individuals of C. sempervirence (The numbers represents the sample codes in Table 4) |
2 Discussion & Conclution
Most conifers have high levels of genetic diversity and low levels of differentiation among populations, as measured by allozymes (Hamrick et al., 1992). However, due to the complex effects of historical factors such as speciation process and Quaternary glaciation, it is difficult to make a priority prediction of the levels of genetic diversity in endangered species (Maki, 2003).
The results of this project represent the first large-scale analysis with nuclear molecular markers to assess genetic diversity of long-lived Cupressus sempervirens trees throughout Iran. The 8 loci analyzed differed greatly in variability level, from loci with few variants for allele to others with abundant polymorphism for alleles widely different in size. All 8 loci were polymorphic, having a total of 113 alleles among the 63 individuals. Previous study conducted on natural stands of Cypress in north of Iran based on the same primers was carried out on 60 different samples. Total number of alleles, mean of effective allele, PIC, average expected heterozygosity (HE) and Shannon indices (HO) was observed in the natural stands of Iranian Cypress (Hafezi et al. 2011) were more than the results of this study. Also average HE and HO were 0.35 and 0.32 in the natural stands of Turkey Cypress (Raddi and Sumer, 1999) and 0.50 and 0.29 in Italian Cypress (Valgimigli et al., 2005), respectively. The analysis shows that Cupressus sempervirence had a low level of heterozygosity. The average Nei’s gene diversity (HE) (0.26) and Shannon indices (HO) (0.41) indicate a low genetic variation within the population. Such a narrow variation can be detrimental to the sustainability of the species. Small populations with low gene diversity will be affected much by lack of gene flow, limited gene drift and reduced mutation rate. Therefore, a high genetic diversity further supports greater differentiation (Nagylaki, 1998; Anand et at., 2004).
Genetic diversity maintained in a plant species is influenced by specific characteristics of the species (Hamrick and Godt, 1989), as well as by its evolutionary history. The low level of genetic variation in C. sempervirence may be attributed to both its evolutionary and cultural history and its very small isolated populations (one to ten individuals) throughout Iran. Cypress trees in Iranian culture were symbol of long life and holly trees in Zoroaster religion (Iranian traditional religion). It seems that is the main reason it̕ s wide distribution and low genetic diversity outside the natural stands (north of Iran) by artificial propagation (seed or cuting) from some long-lived and holly individuals. The identified long-lived Cyprus trees showed very different geographical climatical conditions from humid temprate to semihumid temprate, semi arid cold and arid warm regions in Iran. The clustering and PCo revealed that there are only four group (two big and two very small ones) within population so that the seed or cutting origin of individuals in group 2 were probably Dehbakri 1 (Kerman P.), Harzevil (Gilan P.) and Abarkooh (Yazd P.) and in group 3 were Tange kherghe 6 (Fars P.) and Poshtehan (Gilan P.) because of the oldest trees are attributed to the group and the hypothesis of artifitial propagation.
More genetic diversity resulted in higher cross pollination and higher effective population size. Further, the study implies that more variation needs to be introduced in the existing population for species persistence. Therefore, the immediate approach and need of the hour may be to preserve the local genotypes by collecting seeds from representative ecotypes, introducing more of micropropagated plants with variation to increase the actual population and allow cross breeding.
3 Materials and Methods
3.1 Plant material
The representative plant material was collected from different locations in 14 provinces of Iran (Table 4). Needle samples are collected for 63 individuals. Samples were collected from young branches and middle of crown lengths. They were immediately frozen in liquid nitrogen and then stored in -80℃ before DNA isolation.
 Table 4 Sample code, situation and allometric of studied long-lived trees |
3.2 DNA extraction and primer selection
Total genomic DNA was extracted using the DNeasyTM Plant Mini Kit (QIAGEN) (Valgimigli et al., 2005). DNA was qualified on Ethidium bromide stained Agarose gels and spectrophotometer was used to determine the DNA yield and purity (Doulis et al., 2000). Eight primers were developed by Sebastiani et al. (2005) and Valgimigli et al. (2005) applied them for genetic analysis on C. sempervirens (Table 5).
 Table 5 Characteristics of dinucleotide microsatellite loci developed for C. sempervirense (Sebastiani et al., 2005; Valgimigli et al., 2005) |
3.3 PCR amplification and product detection
The PCR reaction was performed in a final volume of 12.5 μL, containing 25 ng of DNA, 1× reaction buffer, 0.2 mmol/L of each dNTP, 4 mmol/L MgCl2 and 0.8 μmol/L of each primer and 0.75 U Taq polymerase. The following temperature profile was used for amplification: 94℃ for 3 min, then 35 cycles of 94℃ for 1 min, 60℃ for 30 s and 72℃ for 1 min, ending with 72℃ for 5 min. 5 μL of each PCR product were loaded on 1% Agarose gel, run in 1× TAE buffer, stained with Ethidium bromide and photographed under UV light to control the success of the amplifications and to qualify fragments.
The PCR products were labeled with SybrGold fluorescent dye (Invitrogen, Canada) and were loaded on 5% polyacrylamide gels in GS2000 (Corbette Australia). Electrophoresis was performed at 1 200 volt constant power in TBE (1×) buffer as a running buffer, and stopped depending on the real-time dimension of digital image and expected product size of each primer set. Molecular sizes of the amplified fragments were estimated using a 100 bp ladder. Digitally captured image subjected to future analysis.
3.4 Data analysis
Data were scored as presence (1) or absence (0) of a band. Only distinct and well-separated bands were included in the analysis. The polymorphic information content (PIC) was calculated according to Anderson et al. (1993). The Observed and effective number of alleles, Shannon information index (Lewontin, 1972) and Nei's genetic diversity (Nei and Li, 1979) were estimated in POPGEN program (Yeh et al., 1997).
Relationships among individuals were estimated by unweight pair-group mean analysis (UPGMA) by using NTSYS pc 2.02e program (Rohlf, 1998). Cophenetic correlation was used to choose the best clustering method and similarity coefficient. According to cophenetic correlation results, a similarity matrix was generated using Simple Mathing. Principal Coordinate Analysis (PCo) and Genetic distances (Nei, 1972) were conducted by using GenAlex v.6.2. This multivariate approach was chosen to complement the cluster analysis information, because cluster analysis is more sensitive to closely related individuals, whereas PCo is more informative regarding distances among major groups (Hauser and Crovello, 1982).
Acknowledgements
Special thanks to the all foresters in the provinces throughout Iran who helped us to collect the samples.
References
Anand A., Rao C.S., Eganathan P., Kumar N.A., and Swaminathan M.S., 2004, Saving an endangered taxon: Syzygium travancoricum gamble: a case study focusing on its genetic diversity and reintroduction, Physiol. Mol. Bio. Plants, 10: 233-242
Anderson J.A., Churchill G.A., Autrique J.E., Tanksley S.D., and Sorrells M.E., 1993, Optimizing parental selection for genetic linkage maps, Genome, 36: 181-186
http://dx.doi.org/10.1139/g93-024 PMid:18469981
Angielina B., Lorenzo L., Zlatko S., and Raul D., 2011, Variability of wild olives (Olea europaea subsp. europaea var. sylvestris) analyzed by SSR markers and agro-morphological traits, Scientia horticulturace, 129: 561-569
Assadi M., 1998, Cupressaceae in flora of Iran, In: Assadi, M., Khatamsaz, M., Maassoumi A.A., and Mozaffarian V., 21: 8-11, Research Institute of Forests and Rangelands, Tehran: in Persian, pp.8-11
Bilgen B.B., and Kaya N., 2007, Allozyme variations in six natural populations of Scotspine (Pinus sylvestris) in Turkey, Biologia, 62: 697-703
http://dx.doi.org/10.2478/s11756-007-0127-z
Boscherini G., Morgante M., Rossi P., and Vendramin G.G., 1994, Allozyme and chloroplast DNA variation in Italian and Greek populations of Pinus leucodermis, Heredity, 73: 284-290
http://dx.doi.org/10.1038/hdy.1994.135 PMid:7928395
Buiteveld J., Vendramin G.G., Leonardi S., Kamer K., and Geburek T., 2007, Genetic diversity and differentiation in European beech (Fagus sylvatica L.) stands varying in management history, Forest Ecology and Management, 247: 98-106
http://dx.doi.org/10.1016/j.foreco.2007.04.018
Castro V.R., 1998, Chromium in a series of portuguese plants used in the herbal treatment of diabetes, Biol. Trace Elem. Res., 62: 101-106
http://dx.doi.org/10.1007/BF02820025
Chen F., Wang A., Chen K., Wan D., and Liu J., 2009, Genetic diversity and population structure of the endangered and medically important Rheum tanguticum (Polygonaceae) revealed by SSR Markers, Biochemical Systematics and Ecology, 37: 613-621
http://dx.doi.org/10.1016/j.bse.2009.08.004
Darias V., Bravo L., Rabanal R., Sanchezmateo C., Gonzalez Luis R.M., and Hernandez Perez A.M., 1989, New contribution to the ethnopharmacological study of the Canary Island, J. Ethnopharmacol, 25: 77-92
http://dx.doi.org/10.1016/0378-8741(89)90047-0
Doulis A.G., Harfouche A.L., and Aravanopoulos F.A., 2000, Rapid, high quality DNA isolation from cypress (Cupressus sempervirens L.) needles and optimization of the RAPD marker technique, Plant Mol. Biol. Rep., 17(4): 411-412
http://dx.doi.org/10.1023/A:1007679220683
El-Kassaby Y.A., and Benowicz A., 2000, Effects of commercial thinning on genetic, plant species and structural diversity in second-growth Douglas-fir (Pseudotsuga menziesii Mirb Franco) stands, For. Genet., 7: 193-203
Field D., and Wills C., 1996, Long, polymorphic microsatellite in simple microorganisms, Proc. Biol. Sci., 263: 209-215
http://dx.doi.org/10.1098/rspb.1996.0033 PMid:8728984
Giannini R., Morgante M., and Vendramin G.G., 1991, Allozyme variation in Italian populations of Picea abies L, Karst. Silvae Genet., 40: 160-166
Gitzendanner M.A., and Soltis P.S., 2000, Patterns of genetic variations in rare and widespread plant congeners, Am. J. Bot., 87: 783-792
http://dx.doi.org/10.2307/2656886 PMid:10860909
Gupta P.K., Balyan H.S., and Sharma P.C., 1996, Microsatellites in plants: a new class of molecular markers, Current Science, 70: 45-54
Hafezi Shahroodian S., Azadfar D., Soltanloo H., and Ramezanpour S., 2011, Genetic variability in natural Iranian populations of Cupressus sempervirens var. horizontalis in Caspian Sea coastward assessed by SSR markers, Plant Omics Journal, 4(1): 19-24
Hamrick J.L., and Godt M.J.W., 1989, Allozyme diversity in plants. In: Brown A.H.D., Clegg M.T., Kahler A.L., Weir B.S. (Eds.), Plant Population Genetics, Breeding and Genetic Resources. Sinauer, Sunderland, MA, pp.43-63
Hamrick J.L., Godt M.J.W., and Sherman-Broyle S.L., 1992, Factors influencing levels of genetic diversity in woody plant species, New Forest, 5: 95-124
http://dx.doi.org/10.1007/BF00120641
Hauser L.A., and Crovello T.J., 1982, Numerical analysis of genetic relationships in Thelypodieae (Brassicaceae), Syst. Bot., 7: 249-268
http://dx.doi.org/10.2307/2418387
Jarne P., and Lagoda P.J.G., 1996, Microsatellite from molecules to populations and back, Trends in Ecology & Evolution, 11: 424-429
http://dx.doi.org/10.1016/0169-5347(96)10049-5
Jochle W., 1962, Biology and biochemistry of reproduction and contraception, Angew Chem. Weinheim Bergstr Ger., 1: 537-549
PMid:14041949
Lewontin R.C., 1972, The apportionment of Human Diversity, Evol. Biol., 6: 381-398
http://dx.doi.org/10.1007/978-1-4684-9063-3_14
Mahar K.S., Rana T.S., Ranade S.A., and Meena B., 2011, Genetic variability and population structure in Sapindus emarginatus Vahl from India, Gene, 485(1): 32-39
http://dx.doi.org/10.1016/j.gene.2011.05.036 PMid:21723380
Maki M., 2003, Population genetics of threatened wild plants in Japan, J. Plant Res., 116: 169-174
PMid:12736790
Makkonen O., 1967, Ancient forestry, An historical study. Part I. facts and information on trees, Acta. For. Fennica, 82: 1-84
Mascolo N., Autore G., Capasso F., Menghini A., and Fasulo M.P., 1987, Biological screening of Italian medicinal plants for antiinflamatory activity, Phytother Res., 1: 28-31
http://dx.doi.org/10.1002/ptr.2650010107
Nagylaki T., 1998, Fixation indices in subdivided populations, Genetic, 148: 1325-1332
Neel M.C., and Ellstrand N.C., 2003, Conservation of genetic diversity in the endangered plant Eriogonum ovalifolium var. vineum, Conservation Genetics, 4: 337-352
http://dx.doi.org/10.1023/A:1024017029933 http://dx.doi.org/10.1023/A:1024758929728
Nei M., 1972, Genetic distance between populations, Am. Naturalist, 106: 283-292
http://dx.doi.org/10.1086/282771
Nei M., and Li W.H., 1979, Mathematical model for studying genetic variation in terms of restriction endonucleases, Proc. Natl. Acad. Sci., USA, 76: 5269-5273
http://dx.doi.org/10.1073/pnas.76.10.5269
Ponce-Macotela M., Navarro-Alegria I., Martinez-Gordillo M.N., and Alvarez-Chacon R., 1994, In vitro antigiardiasic activity of plant extracts, Rev. Invest. Clin., 46: 343-347
PMid:7839013
Powell W., Morgante M., Andre C., Hanafey M., Vogel J., Tingey S., and Rafalski A., 1996, The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis, Molecular Breeding, 2: 225-238
http://dx.doi.org/10.1007/BF00564200
Raddi S., and Sumer S., 1999, Genetic diversity in natural Cupressus sempervirense L. populations in Turkey, Biochem. Syst. Ecol., 27: 799-814
http://dx.doi.org/10.1016/S0305-1978(99)00028-9
Rehfeldt G.E., Tschebakova N.M., Parfenova Y.L., Wykoff W.R., Kuzmania N.A., and Milyutin L.I., 2002, Intraspecific responses to climate in Pinus sylvestris, Glob. Change Biol., 8: 912-929
http://dx.doi.org/10.1046/j.1365-2486.2002.00516.x
Rohlf F.J., 1998, NTSYS-pc, Numerical taxonomy and multivariate analysis system, version 2.02. Exeter Software, New York, USA
Savolainen O., Pyhajarvi T. and Knurr T., 2007, Gene flow and local adaptation in trees, Ann. Rev. Ecol. Evol. Syst., 38: 595-619
http://dx.doi.org/10.1146/annurev.ecolsys.38.091206.095646
Sebastiani F., Buonamici A., Fineschi S., Racchi M.L., Raddi P., and Vendramin G.G., 2005, Novel polymorphic nuclear microsatellites in C. sempervirens L., Mol. Ecol. Notes, 5: 393-395
http://dx.doi.org/10.1111/j.1471-8286.2005.00938.x
Su Y., Wang T., and Ouyang P., 2009, High genetic differentiation and variation as revealed by ISSR marker in Pseudotaxus chienii (Taxaceae), an old rare conifer endemic to China, Biochemical Systematics and Ecology, 37: 579-588
http://dx.doi.org/10.1016/j.bse.2009.10.005
Toth G., Gaspari Z., and Jurka J., 2000, Microsatellites in different eukaryotic genomes: survey and analysis, Genome Res., 10: 967-981
http://dx.doi.org/10.1101/gr.10.7.967 PMid:10899146 PMCid:310925
Valgimigli M.C., Monaco L.H., Furini A., and La Porta N., 2005, Genetic variability in Italian populations of Cupressus sempervirens L. by SSR and RAPD markers. Dottorato Di Ricerca in Biotecnologie Molecolari Industrialie Ambintali, Universita Degli Studi Di Verona
Vendramin G.G., Michelozzi M., Lelli L., and Tognetti R., 1996, Genetic variation in Abies nebrodensis: a case study for a highly endangered species, For. Genet., 2: 171-175
Wang D.L., Li Z.C., Hao G., Chiang T.Y., and Ge X.J., 2004, Genetic diversity of Calocedrus macrolepis (Cupressaceae) in southwestern China, Biochemical Systematics and Ecology, 32: 797-807
http://dx.doi.org/10.1016/j.bse.2003.12.003
Yeh F.C., Yang R., and Boyle T., 1997, POPGENE: a microsoft windows-based freeware for population genetic analysis: version 1.32, 32 bit, University of Alberta, Edmonton, Canada
Zhang Ch., Chen X., He T., Liu X., Feng T., and Yuan Z., 2007, Genetic structure of Malus sieversii population from xinjiang, China, Revealed by SSR Markers, Genetics and Genomics, 34(10): 947-955
http://dx.doi.org/10.1016/S1673-8527(07)60106-4
. PDF(1690KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Korori A. A. Soudabeh
. Azadfar Davoud
. Shirvany Anoushirvan
. Valipour K. Hossein
. Matinizadeh Mohammad
Related articles
. Cupressus sempervirens
. Long-lived tree
. Genetic diversity
. SSR marker
Tools
. Email to a friend
. Post a comment


